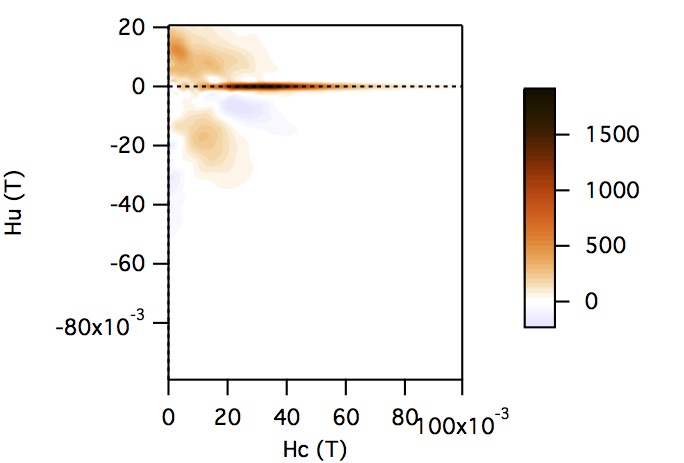
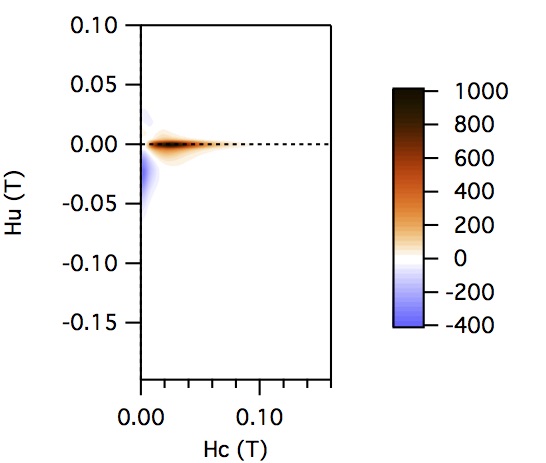
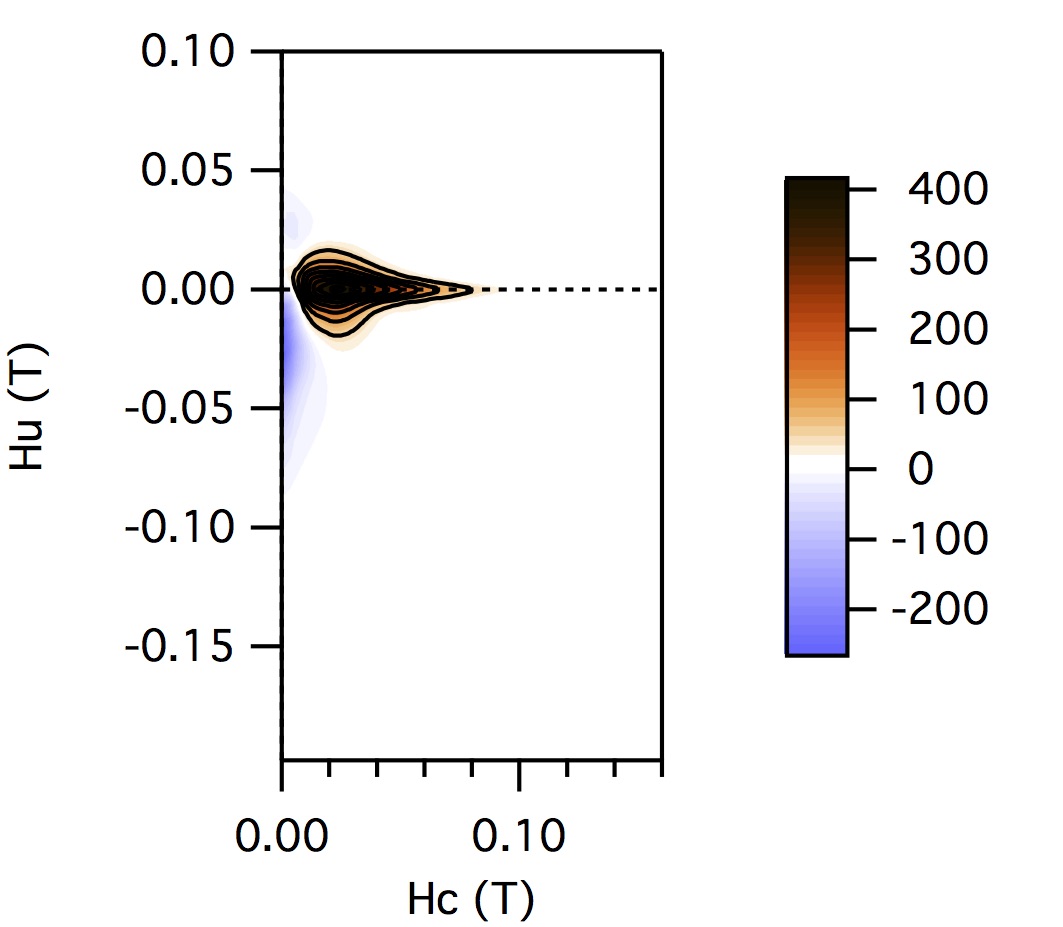
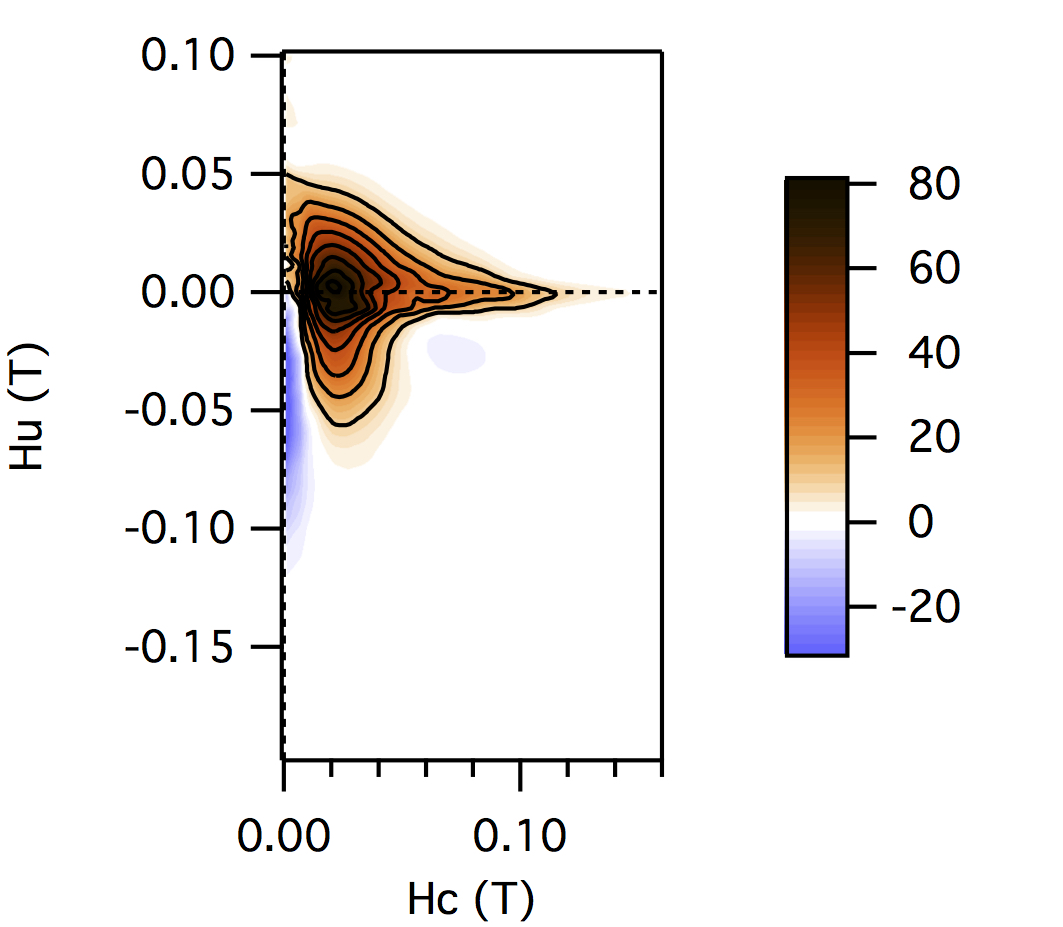
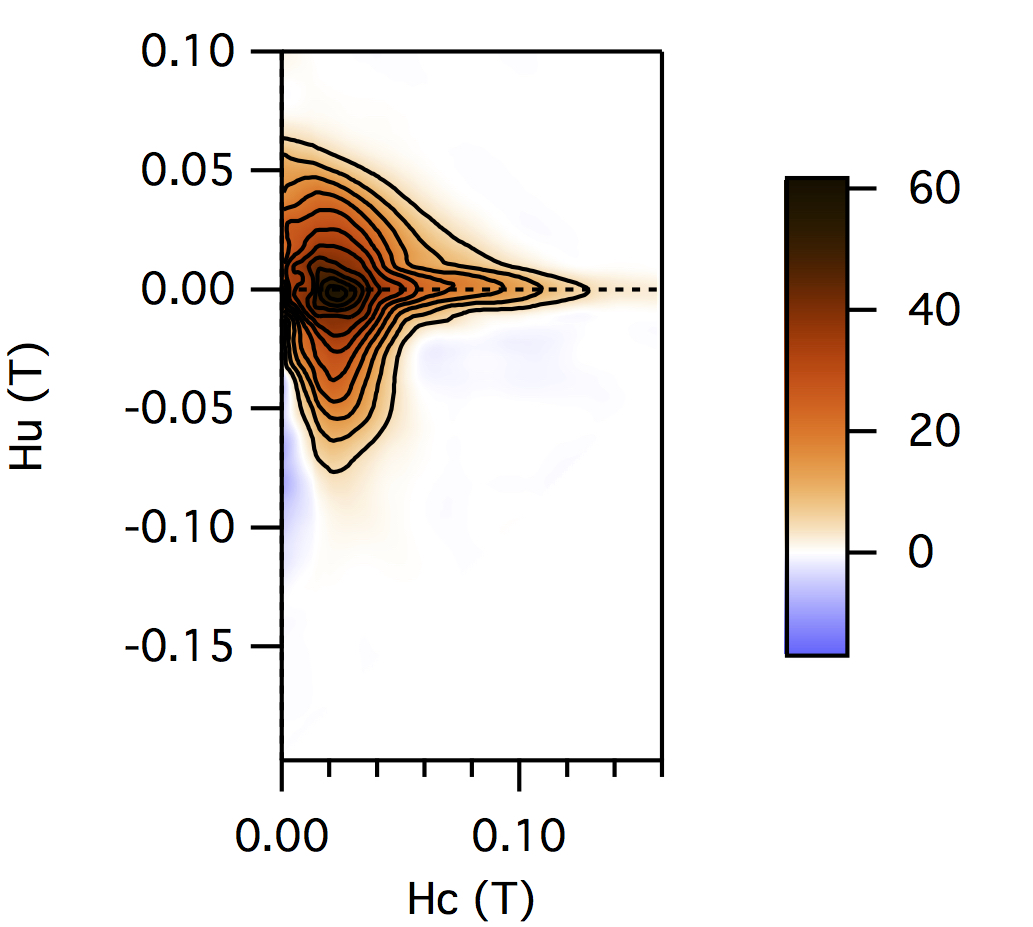
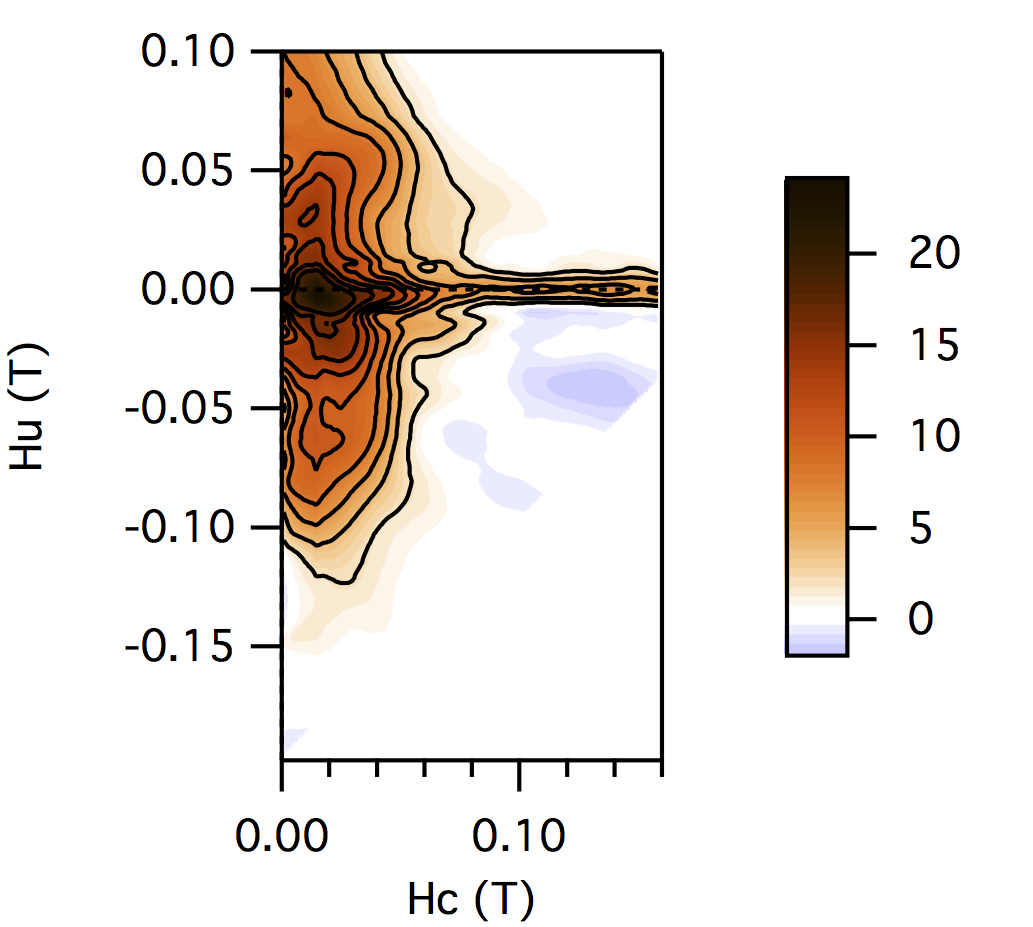
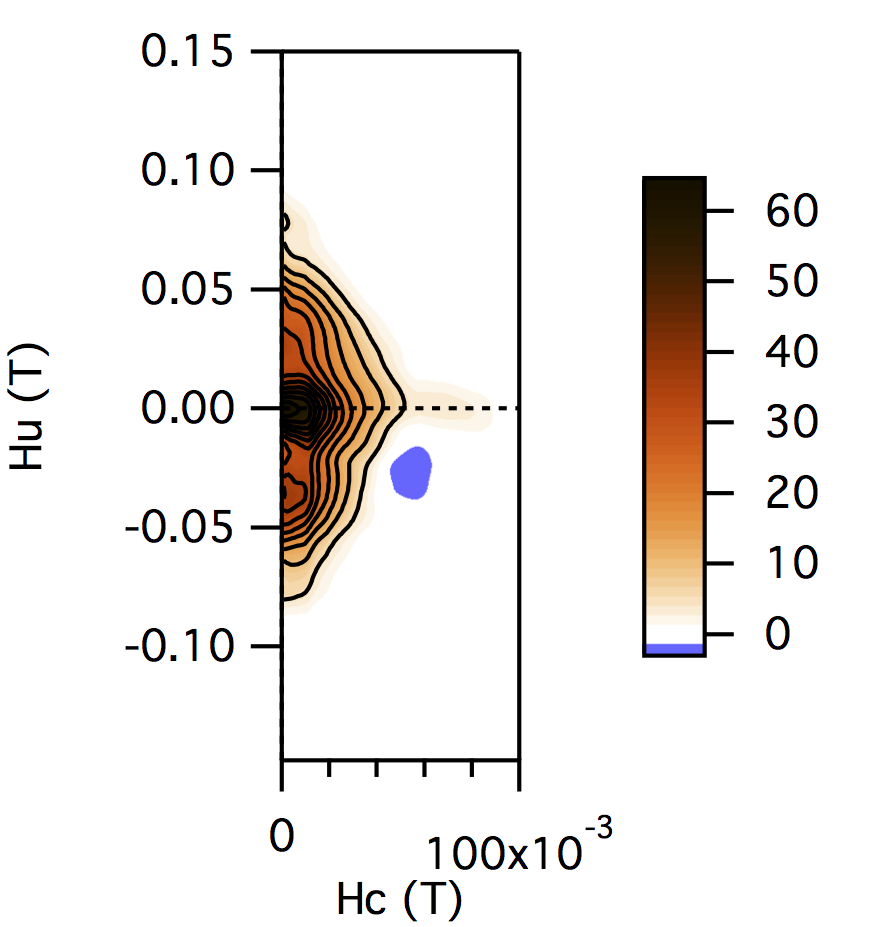
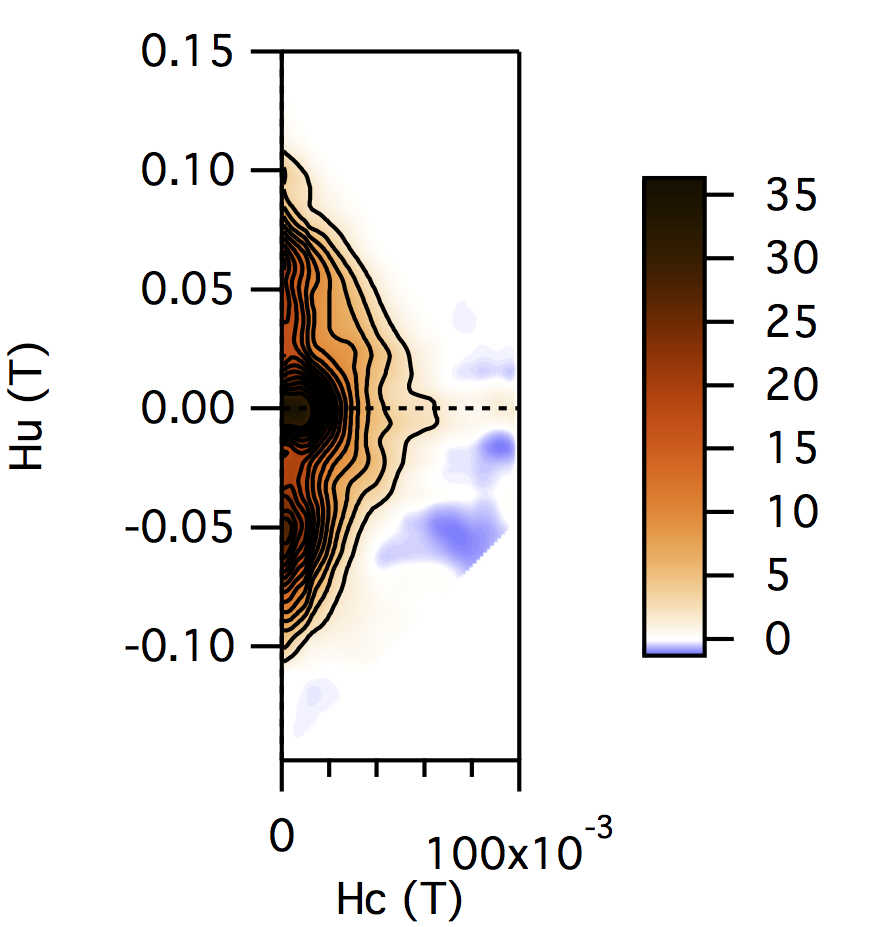
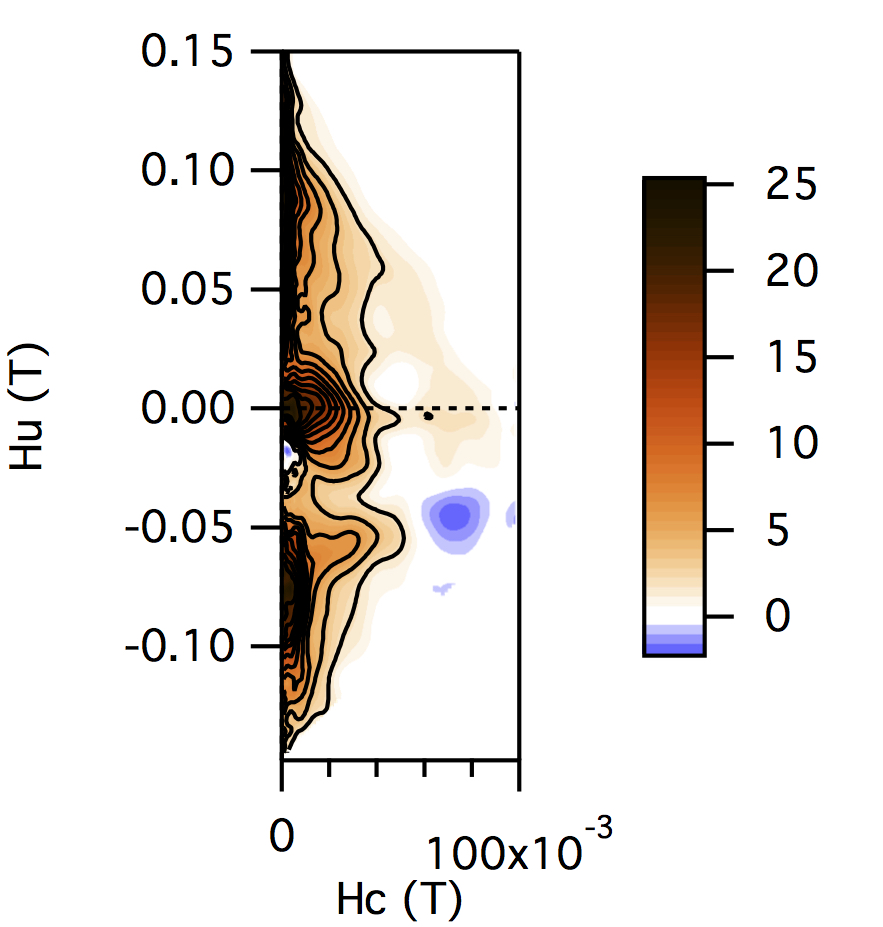
FCC packing of randomly oriented uniaxial magnetite particles
No of Particles: 4
Diameter: 100 nm
Separation of particles: 100 nm
Arrangement: FCC
Easy Axes: Partially aligned along z
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.1 T, Hu =0.02 T, Step Size = 0.0012 T
No. FORCs = 100
No averaging steps: 1000
Lower branch removed: yes
Smoothing factor: 3, 3, 3, 3, 0, 0
No of Particles: 256
Diameter: 100 nm
Separation of particles: 400 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.16 T, Hu =0.1 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 200 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.16 T, Hu =0.1 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 130 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.16 T, Hu =0.1 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 120 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.16 T, Hu =0.1 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 110 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.16 T, Hu =0.1 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 100 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.5 Beta = 20
Simulation Type: LLG
Hc = 0.16 T, Hu =0.1 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 120 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.4 Beta = 40
Simulation Type: LLG
Hc = 0.1 T, Hu =0.15 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 110 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.4 Beta = 40
Simulation Type: LLG
Hc = 0.1 T, Hu =0.15 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes
No of Particles: 256
Diameter: 100 nm
Separation of particles: 100 nm
Arrangement: FCC
Easy Axes: Random
Anisotropy: Uniaxial
Coercivity Distribution: Alpha = 0.4 Beta = 40
Simulation Type: LLG
Hc = 0.1 T, Hu =0.15 T, Step Size = 0.003 T
No. FORCs = 100
No averaging steps: 50
Lower branch removed: yes